What is DNA Barcoding?
What is DNA Barcoding?
DNA barcoding first came to the attention of the scientific community in 2003 when Paul Hebert’s research group at the University of Guelph published a paper titled "Biological identifications through DNA barcodes". In it, they proposed a new system of species identification and discovery using a short section of DNA from a standardized region of the genome. That DNA sequence can be used to identify different species, in the same way a supermarket scanner uses the familiar black stripes of the UPC barcode to identify your purchases.
The gene region that is being used for almost all animal groups, a 648 base-pair region in the mitochondrial cytochrome c oxidase 1 gene (“CO1”), is proving highly effective in identifying birds, butterflies, fish, flies and many other animal groups. The advantage of using COI is that it is short enough to be sequenced quickly and cheaply yet long enough to identify variations among species.
The COI barcode is not effective for identifying plants because it evolves too slowly, but two gene regions in the chloroplast, matK and rbcL, have been approved as the barcode regions for land plants.
The Barcode Production Pipeline
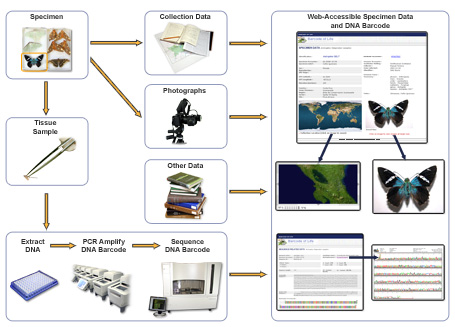
Species identification using DNA barcodes starts with the specimen. Barcoding projects obtain specimens from a variety of sources. Some are collected in the field, others come from the vast collections housed in natural history museums, zoos, botanical gardens and seed banks to name a few.
In the laboratory, technicians use a tiny piece of tissue from the specimen to extract its DNA. The barcode region is isolated, replicated using a process called PCR amplification and then sequenced. The sequence is represented by a series of letters CATG representing the nucleic acids – cytosine, adenine, thymine and guanine.
So if you wrote down the barcode sequence of an Arctic warbler (Phylloscopus borealis), for example, it would look like this:
CCTATACCTAATCTTCGGAGCATGAGCGGGCATGGTAGGC....
And it’s image looks like this:
![]()
Once the barcode sequence has been obtained, it is placed in the Barcode of Life Data Systems (BOLD) database – a reference library of DNA barcodes that can be used to assign identities to unknown specimens.
BOLD is a searchable repository for barcode records, storing specimen data and images as well as sequences and trace files. It provides an identification engine based on the current barcode library and monitors the number of barcode sequence records and species coverage.


